Design of a machine learning approach for bacterial microorganisms detection in metagenomic sequencing without alignment procedures¶
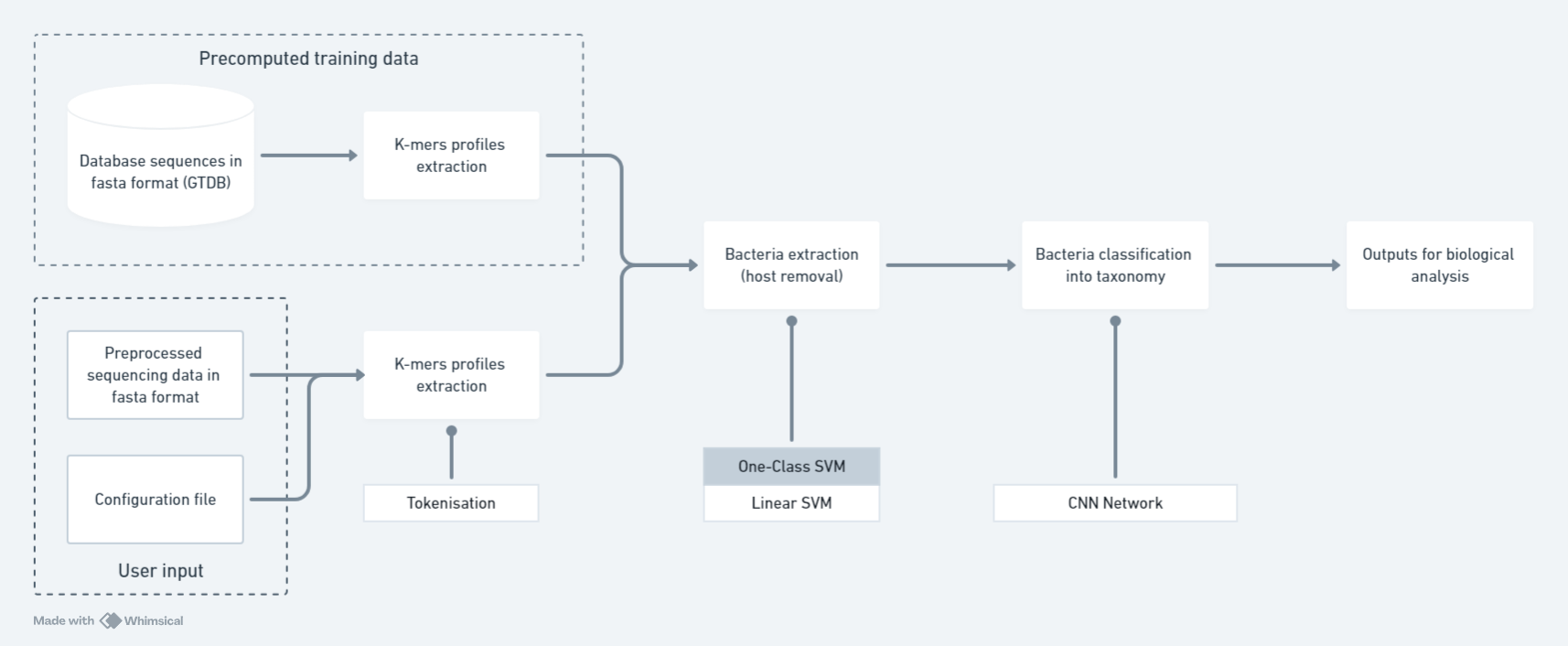
The Caribou pipeline workflow
🎯 Project objective¶
Prior to the project, it was observed that metagenomic sequencing studies often wield results based on a fraction of the data collected from the biological samples. Also, few methods for metagenomic sequencing reads were based on machine learning (ML) methods. Moreover, previous projects in the Diallo laboratory were led to leverage ML methods to classify viral sequencing reads without alignment procedures.
Building on this, the present research project aimed to design and develop a method for classifying bacterial sequencing reads by leveraging ML methods without alignment procedures, and try to valorise a maximum number of reads produced from the original sample. This project was led for my master’s thesis.
📋 Project description¶
To achieve the goal, a pipeline made of four modules was designed.
Data preparation and ingestion
Bacterial reads extraction
Bacterial reads classification
Results output for biological analysis
For both ML steps, multiple models using different methods were used. All models were trained, validated, tested and performances were compared to others trained for the same task in an effort to provide a default method to use in the pipeline as well as optimise the classification performances.
🎨 Design decisions¶
Python 3 programming language
Data ingestion and management using Ray for parallelization and Pandas for subset operations
Machine learning models using Scikit-Learn and Keras
Machine learning parallel training and inferencing using Ray
Synthetic data generation using InSilicoSeq
Computationnaly demanding operations ran on Canadian Numerical Research Alliance clusters
🧾 Key takeaway¶
ML pipeline design
Biological data knowledge
Parallel computing & Ml training