Websites
PowerApp for team process analysis
Héma-Québec
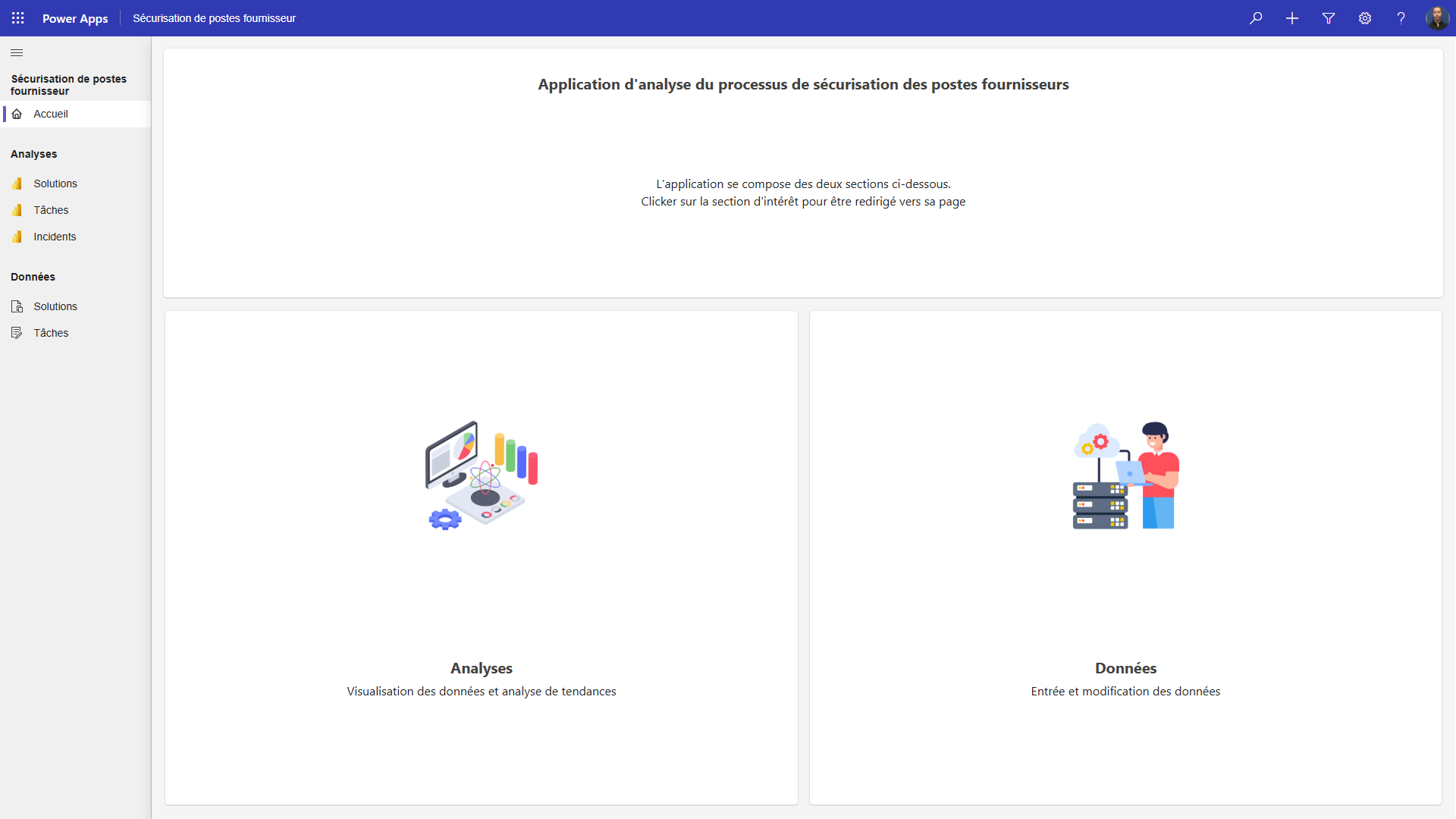
PowerApp for team process analysis - Héma-Québec

Project description:
Model-driven app, used to input, modify, and analyse data relative to a team process. Analysis is then used for improvement of the processes.
Tools & packages used:
- Power Platform Solution
- Power Apps
- Dataverse
- Power BI
- Custom JavaScript & HTML
Responsibilities:
- Front-end & Backend
- Data ETL
- Data Analysis
- Use Case Scenarios with team
Digital portfolio
Personnal project
Digital portfolio - Personnal project
Project description:
This website, I have created this using Github Pages to showcase my work, projects & skills.
Tools & packages used:
- Github Pages
- Jekyll framework
- Liquid templating
- Markdown
- HTML
- CSS
- Javascript
Responsibilities:
- Design & develop pages
- Choose & describe content
- Customize the Jekyll template
WELL-E website v2
DialloLab at UQAM
WELL-E website v2 - DialloLab at UQAM
Project description:
Remake and revamp the WELL-E website in Wordpress so it can be lighter and easier to manage from the backend. Add new features to the website at the same time.
Tools & packages used:
- Wordpress
- Advanced Custom Fields
- Advanced Views Framework
- Blocksy
- GS Teams
- HTML
- CSS
- Javascript
- Twig templating
Responsibilities:
- Recreate previous UI in Wordpress
- Website administration
- Design of Wordpress pages and site (Front end)
- Manage website backend for custom data
- Create custom data formats and UI elements
Other contributors to the project:
- Rachel van Vliet
- Hayda Halmeida
- Mamadou Maladho Barry
DialloLab website
DialloLab at UQAM
DialloLab website - DialloLab at UQAM
Project description:
Remake of the DialloLab website in Wordpress after technical problems with the previous version.
Tools & packages used:
- Wordpress
- Advanced Custom Fields
- Advanced Views Framework
- GS Teams
- HTML
- CSS
- Javascript
- Twig templating
Responsibilities:
- Website administration
- Design of Wordpress pages and site (Front end)
- Manage website backend for custom data
Other contributors to the project:
- Abdoulaye Baniré Diallo (PI)
WELL-E website v1
DialloLab at UQAM
WELL-E website v1 - DialloLab at UQAM
Project description:
Following the end of the contract with ProgramAction, adjust the WELL-E website to ease user experience and adjust UI details
Tools & packages used:
- HTML
- CSS
- Javascript
- JQuery
Responsibilities:
- Adjustments to the website
- Change UI elements
- Facilitate user experience according to communications experts
- Recreate pages
Other contributors to the project:
- Rachel van Vliet
- Catherine Fisette
- Mamadou Maladho Barry
- ProgramAction
- Abdoulaye Baniré Diallo (PI)
PhyML dynamic website implementation
DESS degree project at UQAM
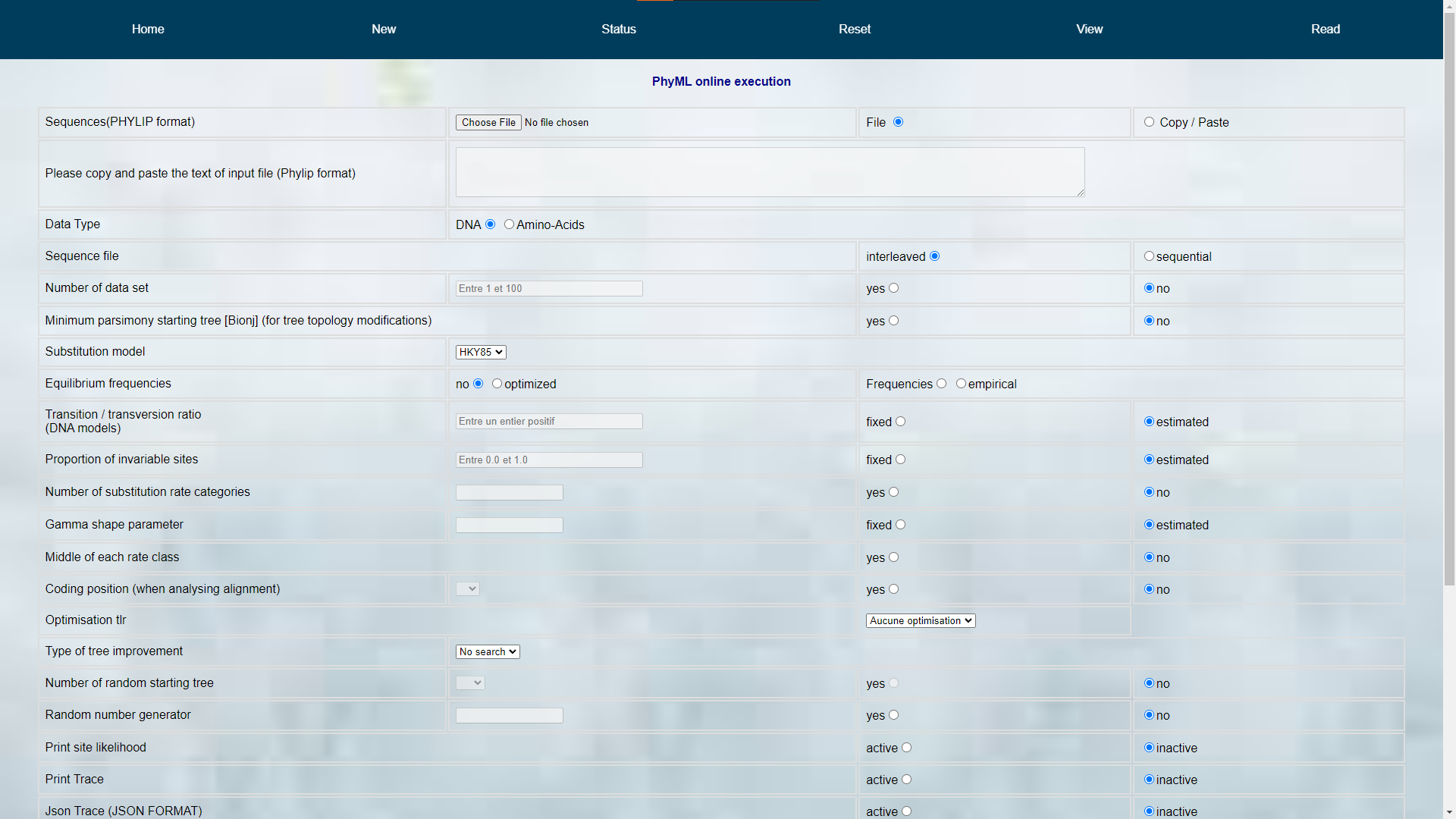
PhyML dynamic website implementation - DESS degree project at UQAM

Project description:
The project consisted in two steps:
- Implement python classes and scripts for launching the PhyML linux CLI precompiled binaries from a local terminal
- Implement a web interface for running the python PhyML python classes previously created
- The frontend interface uses HTML, JavaScript JQuery, and Bootstrap CSS
- The backend leverages a Flask server and an SQLite3 database
Tools & packages used:
- Python
- Javascript
- JQuery
- HTMl
- Bootstrap CSS
- Flask
- SQLite3
- PhyML
Responsibilities:
- Backend development
- PhyML python execution class
- Database python interaction class
Other contributors to the project:
- Abdellatif El Ghizi
- Latifa Mohammadi
- Wanlin Li
RShiny app for data visualization of REAP results
Falcone Lab at IRCM
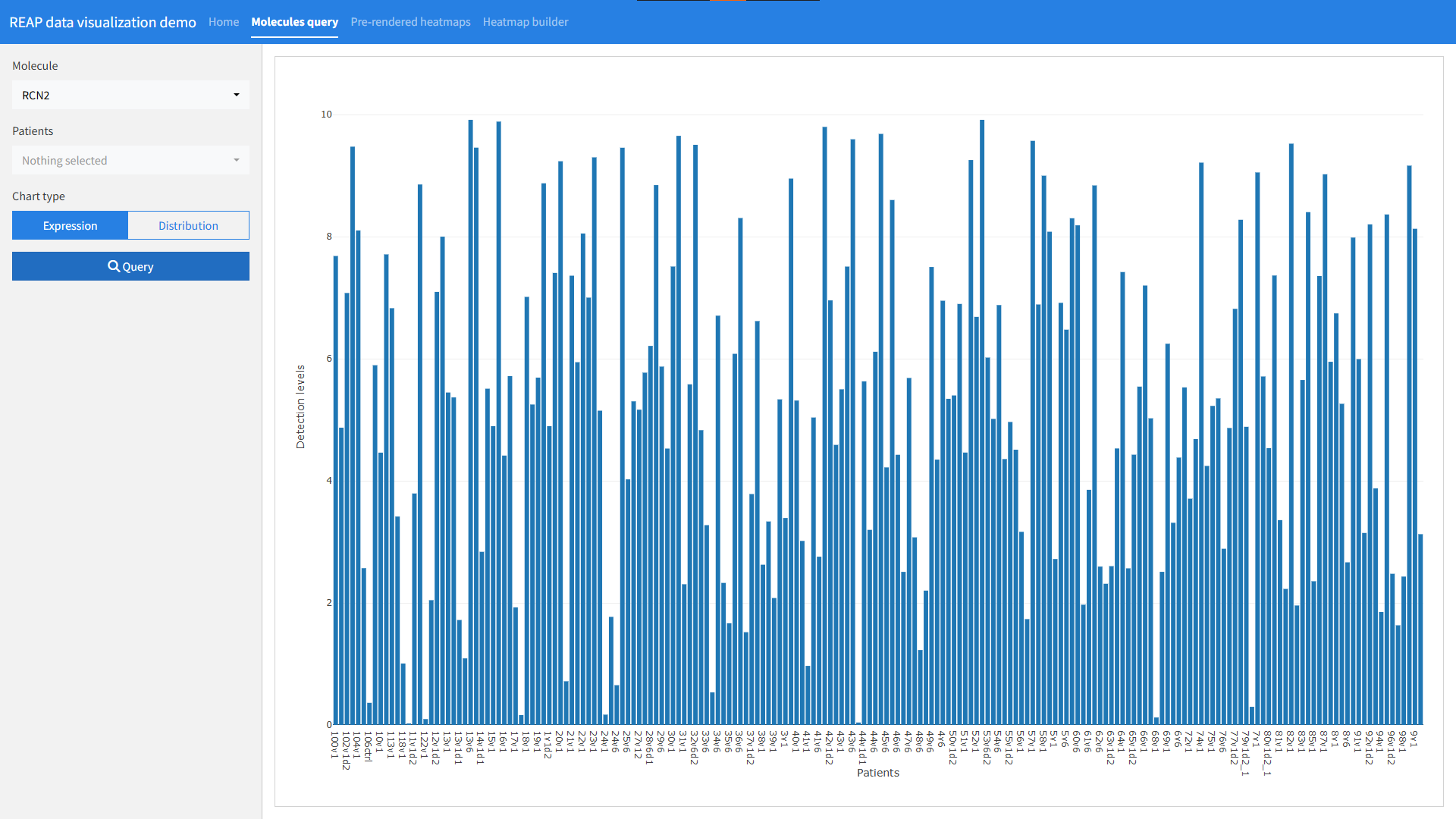
RShiny app for data visualization of REAP results - Falcone Lab at IRCM

Project description:
The project consisted of building an interactive interface for biologists to explore Rapid Extracellular Antigen Profiling (REAP) per patient in the context of long COVID.
The interface allowed biologists to explore the data in three panels.
- Query molecules and visualize either expression per patient of the expression distribution in the whole population or selected individuals group
- Vizualise and explore heatmaps for which antigen groups and patients populations were defined to be of interest by Dr Poudrier
- Build custom heatmaps by selecting antigens and patients from both lists
Tools & packages used:
- R
- plotly
- ggplot2
- RShiny
- R tidyverse
Responsibilities:
- Create original exploration graphs
- Design & develop RShiny application
- Ensure availability of application for users
- Design & add features upon user request
Other contributors to the project:
- Johanne Poudrier (Main user and Biological expert)
 Website
Website
 Code repo
Code repo